Mathematical modelling of dengue fever transmission dynamics in Kenya
Abstract
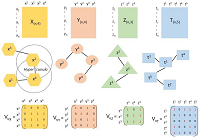
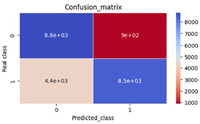
Dengue fever is one of the diseases emerging in Kenya due to effects of climate change and urbanization. The disease is caused by a family of four flavivirus serotypes DENV 1 to DENV4. A deterministic compartmental model for the dengue fever spread dynamics was developed and utilized to examine dengue fever spread dynamics in Kenya. The model was established to be well-stated mathematically and epidemiologically well-posed through positivity and boundedness analysis. The dengue-free equilibrium state was determined as part of the solution to the system of differential equations defining the spread dynamics. The basic reproduction number was determined through the next-generation matrix and used to confirm the stability of the steady state determined before. The study found that when the basic reproduction number was greater than one, the dengue endemic state dominated the solution of the spread dynamics, while when the basic reproduction number was less than one, the dengue free state dominated the solution, implying the disease died down progressively. Sensitivity analysis of the basic reproduction number was carried out to determine the candidate parameters for an optimal control solution. The study found that the infection rate of susceptible mosquitoes, the survival rate of pre-adult mosquitoes, the natural death rate of mosquitoes, the rate at which mosquito survived the extrinsic incubation stage, and the egg-laying of mosquitoes were the most sensitive parameters of the model.
References
[1]Gubler DJ. Dengue, urbanization and globalization: the unholy trinity of the 21st century. Tropical medicine and health. 2011; 39(4SUPPLEMENT): S3–S11,
[2]Khan NT. Etiology of dengue fever. Open Access Journal of Biomedical Science. 2022; 4(2).
[3]Yue Y, Liu X, Ren D, et al. Spatial dynamics of dengue fever in mainland china, 2019. International Journal of Environmental Research and Public Health. 2021: 18(6): 2855.
[4]Guzman MG, Harris E. Dengue. The Lancet. 2015; 385(9966): 453–465.
[5]Wu T, Wu Z, Li YP. Dengue fever and dengue virus in the people’s republic of china. Reviews in Medical Virology. 2022; 32(1): e2245.
[6]Kothai R, Arul B. Dengue fever: An overview. Dengue Fever. 2020.
[7]Cogan JE. Dengue and severe dengue. Available online: http://www. who.int/en/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 2 June 2024).
[8]Medagama A, Dalugama C, Meiyalakan G, et al. Risk factors associated with fatal dengue hemorrhagic fever in adults: A case control study. Canadian Journal of Infectious Diseases and Medical Microbiology. 2020.
[9]Martin E, Chirivella M, Co JKG, et al. Insights into the molecular evolution of dengue virus type 4 in Puerto Ricoover two decades of emergence. Virus Research. 2016; 213: 23–31.
[10]Khetarpal N, Khanna I. Dengue fever: Causes, complications, and vaccine strategies. Journal of immunology research. 2016; 2016: 6803098
[11]Deng SQ, Yang X, Wei Y, et al. A review on dengue vaccine development. Vaccines. 2020; 8(1): 63.
[12]Bosire C, Mutuku F, Ndenga B, et al. A narrative review of dengue fever infection and epidemic activity in Kenya (2010 to 2020). 2023; 12(10).
[13]Konongoi L, Ofula V, Nyunja A, et al. Detection of dengue virus serotypes 1, 2 and 3 in selected regions of Kenya: 2011–2014. Virology Journal. 2016; 13(1): 1–11.
[14]Kamau E, Luka MM, de Laurent ZR, et al. Genome sequences of human coronavirus OC43 and NL63, associated with respiratory infections in Kilifi, Kenya. Microbiology Resource Announcements. 2019; 8(46): e00730–19.
[15]Obonyo M, Fidhow A, Ofula V. Investigation of laboratory confirmed Dengue outbreak in North-eastern Kenya, 2011. PloS One. 2018; 13(6): e0198556.
[16]Attaway DF, Jacobsen KH, Falconer A, et al. Mosquito habitat and dengue risk potential in kenya: Alternative methods to traditional risk mapping techniques. Geospatial Health. 2014; 9(1): 119–130.
[17]Muthanje EM, Kimita G, Nyataya JN, et al. March 2019 dengue fever outbreak at the Kenyan south coast involving dengue virus serotype 3, genotypes III and V. PLOS Global Public Health. 2022; 2(3): e0000122.
[18]Pollett S, Gathii K, Figueroa K, et al. The evolution of dengue-2 viruses in Malindi, Kenya and greater East Africa: Epidemiological and immunological implications. Infection, Genetics and Evolution. 2021; 90: 104617.
[19]Asamoah, JKK, Yankson E, Okyere E, et al. Optimal control and cost-effectiveness analysis for dengue fever model with asymptomatic and partial immune individuals. Results in Physics. 2021; 104919: 31.
[20]Kretzschmar M. Disease modeling for public health: added value, challenges, and institutional constraints. Journal of public health policy. 2020; 41: 39–51.
[21]Khan MA. Dengue infection modeling and its optimal control analysis in East Java, Indonesia. 2021; 7(1): e06023.
[22]Zou L, Chen J, Feng X, Ruan S. Analysis of a dengue model with vertical transmission and application to the 2014 dengue outbreak in Guangdong Province, China. Bulletin of Mathematical Biology. 2018; 80: 2633–2651.
[23]Nyasagare BN, Osman S, Wainaina M. Modelling and analysis of campylobacteriosis in human and animal populations, glob. J. Pure Appl. Math. 2019; 15(5): 551–567.
[24]Onsongo WM, Mwini ED, Nyanaro BN, et al. Stability analysis and modelling the dynamics of psittacosis in human and poultry populations. Commun. Math. Biol. Neurosci. 2022.
[25]Eyaran WE, Osman S, Wainaina M. Modelling and analysis of seir with delay differential equation. Global Journal of Pure and Applied Mathematics. 2019; 15(4): 365–382.
[26]Onsongo WM, Mwini ED, Nyanaro BN, Osman S. The dynamics of psittacosis in human and poultry populations: a mathematical modelling perspective. J. Math. Comput. Sci. 2021; 11(6): 8472–8505.
[27]Zhang T, Dong C, Wang X. Dynamics analysis of a stochastic SIRVI epidemic model with weak immunity. Applied Mathematics Letters. 2024; 149: 108898.
[28]Diekmann O, Heesterbeek JAP, Metz JAJ. On the definition and the computation of the basic reproduction ratio R 0 in models for infectious diseases in heterogeneous populations. Journal of Mathematical Biology. 1990; 28: 365–382.
[29]Van den Driessche P, Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Mathematical Biosciences. 2002; 180(1-2): 29–48.
[30]Muia DW, Osman S, Wainaina M. Modelling and analysis of trypanosomiasis transmission mechanism. Global Journal of Pure and Applied Mathematics. 2018; 14(10): 1311–1331.
[31]Osman S, Otoo D, Sebil C. Analysis of listeriosis transmission dynamics with optimal control. Applied Mathematics. 2020; 11(7): 712–737.
[32]Osman S, Otoo D, Daniel Makinde O. Modeling anthrax with optimal control and cost effectiveness analysis. Applied Mathematics. 2020; 11(3): 255–275.
[33]Ito H. Input-to-state stability and Lyapunov functions with explicit domains for SIR model of infectious diseases. Discrete and Continuous Dynamical Systems-Series B. 2021; 26: 9.
[34]Osman S, Daniel Makinde O. A mathematical model for coinfection of listeriosis and anthrax diseases. International Journal of Mathematics and Mathematical Sciences. 2018; 1: 1725671.
[35]Theuri DM. Stability analysis and modelling of listeriosis dynamics in human and animal populations. Global Journal of Pure and Applied MatheMatics. 2018; 14(1): 115–138.
[36]Anthony Eustace K, Osman S, Wainaina M. Mathematical modelling and analysis of the dynamics of cholera. Global Journal of Pure and Applied Mathematics. 2018; 14(9): 1259–1275
Copyright (c) 2024 Brian Nyanaro, George Kimathi, Mary Wainaina

This work is licensed under a Creative Commons Attribution 4.0 International License.